这是一个美国的Matlab代写,主要与肌电信号分析相关
Please answer all questions thoroughly, using charts and/or explanations as requested and where appropriate. In addition
to the lab report, you must also turn in your MATLAB code to receive full credit. The code should be able to run from
beginning to end without any interventions. I have included labAnalysisCodeTemplate.m to help you get started.
• Load your data (i.e, Group1_1.mat, etc…) into MATLAB using the load command. Type held load if you need help with
the syntax. Do not just load your code into the command window. Actually put the command into your script.
Remember that your final submitted script needs to be able to do everything without any intervention. Your data file
will contain three structured variables: MVC, CURLS, and FATIGUE.
Part 1: RMS Filtering and Maximum Voluntary Contractions
MVC is a k element cell array where each element contains fields for each MVC subject and trial. For example, MVC{1}
may contain data for biceps MVC (or another EMG, depending on the order you placed the EMG electrodes). Note the use
of { }, rather than ( ). Each MVC{i} contains the task name (‘MVC_xxx’), raw EMG data, time vector, and raw accelerometer
(ACC) data. The EMG data is time x 4, where the column corresponds to the sensor of interest (biceps, triceps, FDS, EDC
EMG sensors).

The figure above shows a block diagram of how to convert a recorded EMG signal into a smoothed DC envelope. The
envelope magnitude is what gives you the relative force of the EMG. Your data collected in the lab represents the
Amplified EMG. You will need to determine the Smoothed DC Voltage envelope.
I have written an rms.m function that will take as input an EMG signal and it’s time vector and output a smoothed DC
envelope. The syntax is smoothedSignal = rms(emgVector, timeVector, windowLength, overLap). Type help rms to see
what each of these variables represent.
You will investigate how the windowLength and overLap affect the smoothed DC envelop. Remember that the rms is
calculated on the rectified signal, not the original signal.
• Create a 4×1 figure (1 figure for each subject in your group), and on each subplot, plot in black the raw BICEPS emg
recorded during the BICEPS MVC task. Then over each subplot, plot in red the smoothed envelope calculated using a
A) windowLength=50 samples, overLap=5 samples, B) windowLength=50 samples, overLap=40 samples, C)
windowLength=400 samples, overLap=40 samples, and D) windowLength=400 samples, overLap=320 samples. Make
sure you properly label your subplots with axis labels and an informative title (e.g. ‘Envelope calculated using
windowLength=400, overLap=320’).
• Put this figure into your lab report. Insightfully comment on the effect of changing the values of the windowLength
and overLap. What does the rms function do to the signal? From the values you just examined, choose (and state in
your report) values for windowLength and overLap that you will use for the remainder of your analyses.
• On a second 4×1 figure (again, 1 for each subject), plot the EMG from the MVC trials of each of the four muscles. In
other words, plot the biceps EMG for the biceps MVC trial, plot the triceps EMG for the triceps MVC trial, and so forth.
Like in the previous figure, plot first the raw EMG data in black, and then the smoothed DC envelop in red, calculated
using your chosen parameters. Label your plots appropriately.
• Find and report the maximum value of the smoothed envelope for each muscle’s MVC. Comment on whether (and
why) these maximum values for the RMS of each muscle do (or do not) make sense, relative to each other.
Part 3: Relationship between EMG and Force
Here you will determine the relationship between Force and EMG envelope magnitude during the bicep curls. The CURLS
variable contains the same fields as the MVC variable, with an additional weight field which tells what weight was curled.
This field is initially set to NaN. You will need to change it in your code to the weights that you used.
• Make a series of subplots that show for each curl set (i.e. at each weight) 1) the raw and smoothed DC envelope EMG
signals, and 2) immediately below that the Z-component of accelerometer data from the FDS electrode. You should
smooth the accelerometer data using a low pass filter with a 2Hz cutoff (all data has about a 2kHz sampling rate). An
example of the subplots you should create is shown below for 1 curled weight. You should make similar plots for all
curled weights. Raw signals are in black, and smoothed signals are in red.
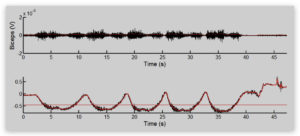
• From the accelerometer plots, determine a threshold level (red line in above figure) such that you will calculate the
average EMG activity when the accelerometer value is below that threshold. Your threshold should be appropriate
for all weights used. For example, in the above figure, I could choose -0.44, so that I will find the average EMG only
during those specific portions of the cur when the accelerometer data is less than the threshold. Note again, that you
should choose a single threshold for ALL weights. Report your accelerometer threshold.
• Find the average EMG activity during each curl (according to your chosen threshold). Do this for each weight. Thus,
assuming you did four weights and ten curls at each weight, you will end up with a 10×4 (curls by weight) matrix.
• Plot the mean and standard deviations of the EMG vs curled weight. Consider using the built-in function errorbar.
• Discuss the relationship you observed between EMG and force. Discuss any deviations from linearity.
• Properly annotate all of your plots. Make sure you have plots for all subjects in your group.


